Question: How does Chlamydia do so much with so little?
Overview
The Ouellette lab is interested in the consequences of reductive evolution on bacterial physiology. As Chlamydia has adapted to an intracellular niche, it has lost many genes that are present in free-living bacteria. These genes/gene pathways that have been lost through reductive evolution (Muller’s Ratchet) typically encode metabolic pathways to synthesize, for example, amino acids. Since Chlamydia relies on its host cell for many nutrients, it is not surprising that it would eliminate these types of genes. However, Chlamydia has also eliminated genes that are considered essential for viability or pathogenesis in many other bacteria. This raises many interesting questions that are the focus of Dr. Ouellette’s research as described below. Conversely, when Chlamydia has retained genes that are atypical for Gram-negative bacteria (e.g. genes normally found in Gram-positive bacteria), this is also interesting and suggests a function that is important to chlamydial growth - otherwise these genes would have been deleted.
Chlamydial Cell Division
Chlamydia is amongst the rare bacteria that lack critical cell division proteins and how these organisms manage to divide in their absence is an intriguing and unanswered question. The cell division apparatus in bacteria forms a supra-molecular interaction complex of multiple proteins that ultimately separates two equally-sized daughter cells in a process called binary fission. Canonically, the building of this complex proceeds in a step-wise fashion such that early proteins recruit later ones. Where Chlamydia is unusual is that it has no homologues of the early proteins, in particular FtsZ, which is considered the central organizer of the division apparatus.
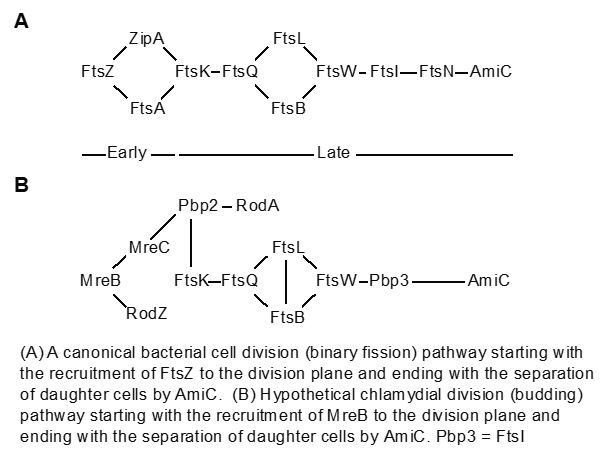
We demonstrated that Chlamydia uses an alternate protein to substitute for FtsZ (Ouellette et al., 2012; Lee et al., 2020). This protein, MreB, has similar characteristics to FtsZ, can form a ring at the septum, and can interact with other cell division proteins, in part mediated by a unique N-terminal domain. Work with collaborators from UTHSC in Memphis indicated that chlamydiae divide by a budding mechanism rather than by binary fission (AbdelRahman et al., 2016; Ouellette et al., 2020). This represents an exciting finding that we continue to study, including how division is inhibited during different stress states (see below in Chlamydial Persistence). We hypothesize that Chlamydia uses an MreB-dependent polarized division mechanism. Given that Chlamydia is a coccoid bacterium, we are interested in understanding how its septum is formed at a specific site on one side of the cell. Current project goals are:
- to determine how the chlamydial septum is localized to a specific site on one side of the mother cell,(Ouellette et al., 2022),
- to understand the role of phospholipids in constructing the budding daughter cell and factors that regulate phospholipid synthase activity/localization,
- to identify novel chlamydial division proteins and regulators that interact with MreB using a system for monitoring protein-protein interactions in bacteria.
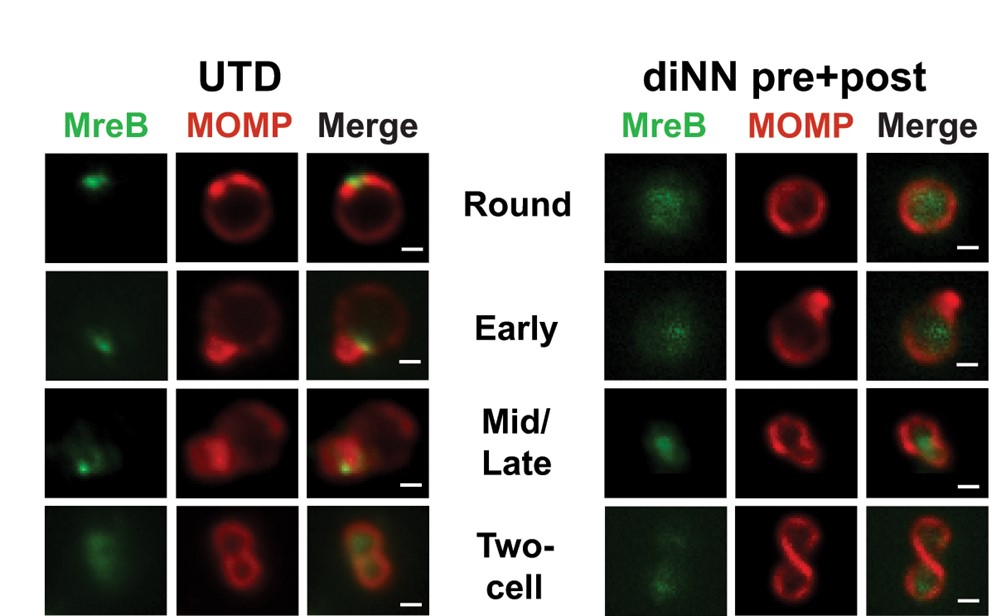
The localization of MreB is shown in untreated (UTD) chlamydiae at different stages of
division. Note its localization to one side initially and then to the septum between the
daughter and mother cell. Treating with a drug (diNN) to disrupt cardiolipin delocalizes
MreB from the membrane. (See Ouellette et al, 2022).
Most recently, we identified and characterized a novel cytoskeletal protein involved in chlamydial cell size and are investigating its function (Brockett, Lee et al., 2021). A follow-up study has characterized the unique N-terminal domain of BacA, and we have performed assays to identify potential interaction partners of this protein, which are under investigation. Other projects under investigation include how cell division is regulated over the course of the developmental cycle. We currently employ a variety of molecular, biochemical, and cell biology techniques for these projects and investigate the proteins of interest involved in these systems. A better understanding of chlamydial cell division and size regulation may lead to the identification of novel therapeutic targets that would eliminate Chlamydia specifically without disrupting normal flora. This project is a collaboration with the Cox Lab at the University of Tennessee Health Science Center in Memphis.
Chlamydial Persistence
While productive growth through the normal developmental cycle results in the generation of EBs, non-productive growth leads to the establishment of persistent forms of the organism. Persistent chlamydiae are most readily identified by their aberrant morphology: they resemble enlarged RBs that fail to undergo cell division.
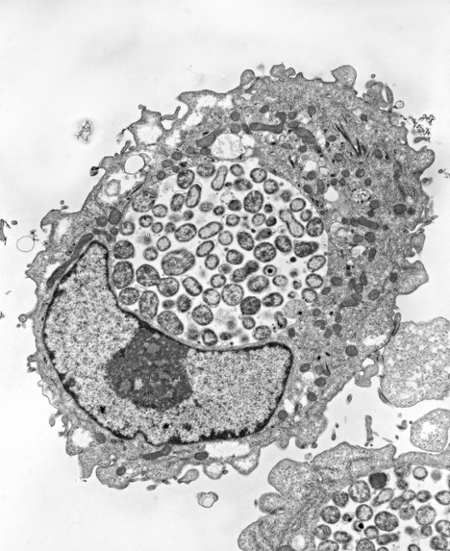
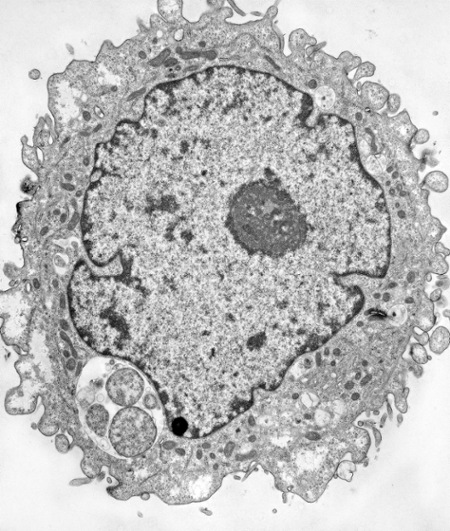
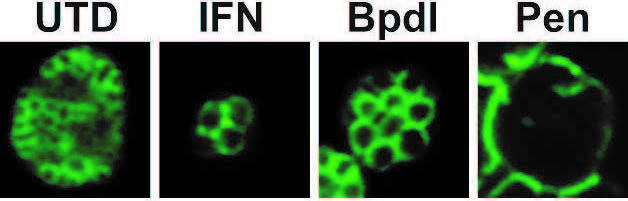
Electron micrographs of HEp-2 cells infected with Chlamydia pneumoniae and grown for 48hr in the absence (left) or presence (right) of IFN-gamma. The nucleus has a black border whereas the inclusions have a white background with grey circular shapes in them. The inclusion in the IFN-gamma treated cell is smaller with fewer organisms that are enlarged compared to the untreated infected cell that has dozens of RBs. From Ouellette et al., 2006.
Induction of persistence is a reversible process defined by viable, culture-negative growth that does not result in the production of infectious EBs. This requires a long-term association with the host cell. Chronic sequelae associated with chlamydial infection may be caused by a persistent form of the organism. A better understanding of the mechanisms chlamydiae use to maintain a persistent growth state will lead to improved diagnostic and therapeutic strategies. Host immune effectors, b-lactam antibiotic treatment, and nutrient deprivation can induce persistence in vitro. The mechanisms by which each of these elicits persistence differ. In humans, the immune cytokine IFN-gamma activates target cells to produce indoleamine-2,3-dioxygenase (IDO). IDO decyclizes tryptophan to N-formyl-kynurenine, which results in an enzymatically controlled tryptophan-limiting environment. Because chlamydiae are dependent on the host cell for tryptophan, the bacteria are effectively starved for this essential amino acid. Penicillin and other b-lactam antibiotics have the phenotypic effect of blocking cell division, presumably by interfering with the action of penicillin-binding proteins in this process. Starving chlamydiae for essential nutrients such as vitamins and iron can also induce a persistent growth state. A critical parameter of persistence is that it is reversible: the organism remains viable such that it can re-enter the productive growth cycle once the inducing stress has been removed. There are data to suggest that persistent chlamydiae are refractory to clinical antibiotic treatment, which may also explain treatment failures.
Most bacteria respond to amino acid limitation by engaging a stringent response, which is a transcriptional program used to adapt to nutrient-poor conditions. Interestingly, the stringent response is also necessary for inducing persister cells, which survive stressful conditions (such as antibiotic treatment) without becoming genetically resistant to the stress. Chlamydia lacks the genes necessary for implementing a stringent response but are capable of persisting. Additionally, Chlamydia has very few identified regulatory elements thus, how it responds to stress is an intriguing question. We have previously shown (Ouellette et al., 2006) that Chlamydia has an unusual response to IDO-mediated tryptophan limitation: it globally increases transcription while translation is globally decreased. This disconnect between transcription and translation is unusual in bacteria. Our goals are to understand how and why Chlamydia responds in this way. Recent work has shown that both Chlamydia and Streptococcus pyogenes (another Trp auxotroph) respond similarly to Trp starvation (Ouellette et al., 2021). We hypothesize that ribosome stalling at Trp codon-rich sequences negatively impacts transcription and translation, resulting in the phenotypes observed. Current efforts are designed to mechanistically understand this observation as well as the consequences to the bacterium when this occurs. Having characterized the ability of tRNA synthetase inhibitors to phenocopy the effects of IFN-gamma (Hatch and Ouellette, 2020), we will use these tools along with CRISPRi to investigate the genetic mechanisms that control this response in bacteria with a variety of transcriptional techniques ranging from traditional (e.g. Northern blots and qPCR) to cutting-edge (e.g. RNAseq, ChIP-seq, and ribosome profiling). Results will lead to the identification of novel therapeutic and diagnostic targets that have the potential to identify and treat asymptomatic chlamydial infections. This project is funded by a CAREER award from the National Science Foundation. Other persistence related projects supported by a multi-PI R01 from the NIAID/NIH with Drs. Carabeo and Rucks focus on how chlamydial cell division is inhibited during persistence (see Riffaud et al., 2023).
Differentiation through Degradation
As Chlamydia differentiates from one developmental stage to the other (i.e. RB to EB or vice versa), they must necessarily reorganize their protein content between these morphologically and functionally distinct forms – this requires the action of proteases. We hypothesize that protease activity is essential for chlamydial differentiation. More specifically, the regulated degradation of key targets drives differentiation of RBs to EBs. Chlamydia has a number of conserved protease systems, and we are investigating several of them for their function in chlamydial growth and differentiation.
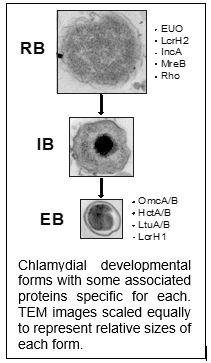
Of particular interest is the Clp protease system, composed of a proteolytic subunit, ClpP, and a substrate recognition subunit, ClpX or ClpC, that identifies and unfolds proteins into ClpP for degradation. ClpP is a validated drug target in various bacteria, including Chlamydia (Seleem et al., 2020). Unusually, Chlamydia possesses two ClpP isoforms, and we have shown that they have differential effects in Chlamydia (Wood et al., 2018). Interestingly, disrupting the activity of ClpX results in non-functional EBs (Wood et al., 2020). Recent work from our group has shown that the ability of ClpX to switch substrate specificity may govern the transition between RBs and EBs (Wood et al., 2022). Current work is focused on (i) identifying substrates of ClpX as well as the critical domains and residues important for its function and (ii) characterizing the function of the related unfoldase ClpC and its substrate preferences. This work is currently supported by an R01 award from the NIAID/NIH and is a collaboration with the Fisher Lab at Southern Illinois University at Carbondale.
We have identified a novel transcriptional regulatory circuit and are investigating its effects. With the recent development of genetic tools for Chlamydia, the Ouellette Lab is also taking advantage of these advances by developing inducible repression systems based on CRISPR interference to knockdown gene expression in Chlamydia (Ouellette et al., 2021). We continue to develop new tools and plasmids for use in Chlamydia to help us study essential systems.
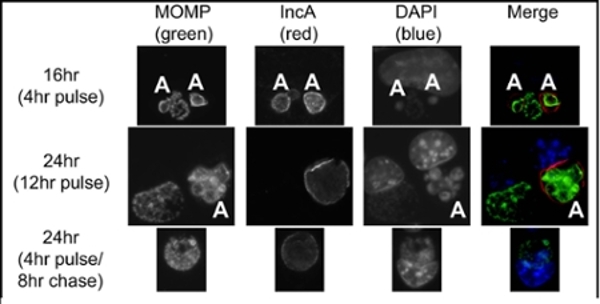
Test of CRISPRi system in Chlamydia. Cells were infected with a mix of inducible CRISPRi transformants (target IncA) and wild-type Chlamydia in the presence of penicillin (selective antibiotic for the plasmid). Samples were induced with anyhydrotetracycline (aTc) at 12 hr post-infection (hpi) and assessed at 16 and 24 hpi, as indicated. Pulse indicates that aTc was present from 12hpi until fixation, whereas pulse/chase indicates aTc was washed out at 16hpi, then fixed at 24hpi. MOMP=chlamydial major outer membrane protein. IncA is the target of the system. DAPI stains DNA. Aberrant Chlamydia (those without the CRISPRi plasmid) are indicated with ‘A’ and still express IncA, while Chlamydia expressing the CRISPRi construct do not express IncA, unless aTc is removed (4hr pulse/8hr chase).
Wolbachia in Aquatic Insects
In collaboration with Dr. Jeff Wesner (http://jeffwesner.wordpress.com/) in the Biology Department at the University of South Dakota, we are investigating the presence of another obligate intracellular bacteria, Wolbachia, in insects from the Missouri river tributary systems around Vermillion, SD. Our initial analyses have revealed that Wolbachia infects a number of aquatic insect species (Sazama et al., 2017, 2019), and we are currently trying to determine what effect this has on the insect and how Wolbachia is transferred between individuals of a species and between species.

