Peter C. Iwen, PhD
Research activities in my lab include the development and optimization of molecular assays for the identification of microbial pathogens with an emphasis on fungi and bacteria (including the Mycobacterium species).
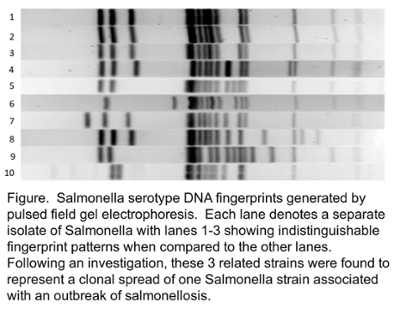
Original work in my laboratory showed that the internal transcribe spacer region within the rDNA complex was a useful target for the identification of both the common and uncommon fungal and mycobacterial species. Subsequent work has been in utilization of whole genome sequencing in collaboration with bioinformaticians to analyze microbial pathogens to investigate foodborne and nosocomial outbreaks as well as to monitor for genetic targets to identify the presence antimicrobial resistance.
Prabagaran (Praba) Narayanasamy, PhD
The Narayanasamy Laboratory is working on Mycobacterium, HIV virus, MRSA, Pseudomonas aeruginosa and Acinetobacter baumannii. Over 4 billion people are infected by some kind of pathogens and are more likely to develop into active infection. Gram-negative bacteria have intrinsic abilities to develop new mechanisms of resistance and can pass along genetic materials that allow the next generation of bacteria to become drug-resistant as well. Thus, there is a critical need for new antibiotics and drug delivery techniques to meet the needs of patients now and in the future. Our lab is working on asymmetric synthesis of drug compounds, drug discovery against drug resistant pathogens targeting MEP pathway, menaquinone pathway and iron metabolism. The property of cyclic diphosphate is also studied thoroughly. In addition we also use nanoparticle technology to target drug resistant pathogens and co-infection in macrophage. In addition we also discovered that Ga nanoparticle could be used as a single drug to treat HIV-TB co-infection. We will be optimizing the Ga nanoparticle to determine best results by in vitro and in vivo. Ultimately, these data could lead to the discovery of new drugs or techniques to treat drug resistant infections, with the potential that other human pathogens could be susceptible as well. In addition we also work in neurons development. Methyl glyoxal, a toxic material generated by glycolysis in brain, affects the survival of neurons. To protect the neurons we enhance the glyoxalase pathway to detoxify the methyl glyoxal. We also design, synthesize and test novel compounds to enhance glyoxalase pathway for detoxification. The successful flavonol antioxidants from our preliminary studies in cerebellar neurons will be tested for reducing aging and Alzheimer’s disease in animal models.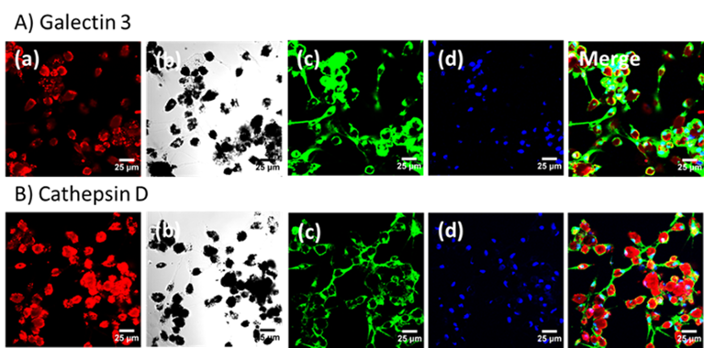 (photo by Narayanasamy Lab. Confocal images of macrophages treated with GaNP. a) Fluorescein-conjugated GaNP (Red), b) macrophages and GaNP, c) galectin 3 (A) or cathepsin D (B), d) Nuclei (Blue). Nanoparticle-treated MDMs were incubated with primary rabbit galectin 3 or cathepsin D. These proteins were visualized by incubation with Alexa Fluor®488 goat-anti-rabbit antibody. The scale bar represents 25 μm.)
(photo by Narayanasamy Lab. Confocal images of macrophages treated with GaNP. a) Fluorescein-conjugated GaNP (Red), b) macrophages and GaNP, c) galectin 3 (A) or cathepsin D (B), d) Nuclei (Blue). Nanoparticle-treated MDMs were incubated with primary rabbit galectin 3 or cathepsin D. These proteins were visualized by incubation with Alexa Fluor®488 goat-anti-rabbit antibody. The scale bar represents 25 μm.)
Thomas McDonald, PhD
Our laboratory conducts research related to acute phase proteins and their role in the inflammatory process. Acute phase proteins are produced by the liver in response to proinflammatory cytokine stimulation, primarily interleukin (IL-) 1, IL-6, and tumor necrosis factor alpha (TNFα). Our laboratory has focused on the acute phase protein serum amyloid A (SAA). Within 18 hours following stimulation, SAA is produced by the liver and blood levels reach 1000 fold higher concentrations than levels prior to stimulation. The function of SAA is unknown, although it plays a role in lipoprotein metabolism. Our laboratory has discovered a novel isoform of SAA that is produced extrahepatic and does not occur in the circulation. It was discovered in bovine colostrum and was unique to that fluid in that it was not present in milk five days following parturition. Moreover, a unique N-terminal amino acid sequence found in this bovine SAA isoform was identical for SAA isolated from the colostrum of five different animal species. As with bovine, the new SAA isoform only associated with colostrum and not milk or serum. We have prepared a variety of recombinant proteins and peptides that reflect the novel colostrum-associated amino acid sequence, and have determined that one functional role of this unique isoform of SAA causes increased mucin-3 production by intestinal cells. Mucin-3 is essential for protecting the gut from bucteral adherence, colonization and infection. Based on either diagnostic assays of secreted biological fluids for detection of infection and inflammatory condition or of serum amyloid a isoform from colostrum, we have had twelve issued patents and four pending. These and other findings from our laboratory imply that this heavily conserved amino acid region of the colostrum-associated SAA molecule has an important beneficial function in the health and well being of the neonate, in particular with respect to the protection of the GI tract from pathogens encountered early in life. I have invented, developed and manufactured prototype diagnostic tests for human and veterinary applications as well as established start-up companies to manufacture and market these technologies in collaboration with companies worldwide.
Guangshun (Gus) Wang, PhD
My research focuses on the identification, characterization, and engineering of novel antimicrobial agents based on structural, bioinformatics, and functional studies. Our ultimate goal is to develop novel compounds that curb pathogenic microbes, especially difficult-to-kill microbes such as methicillin-resistant Staphylococcus aureus (MRSA).
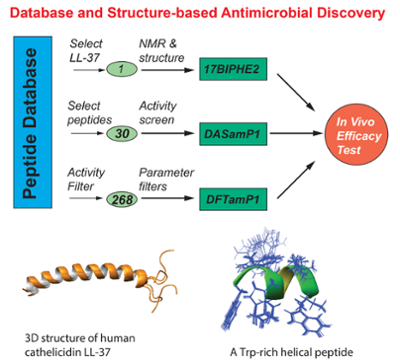
Naturally occurring antimicrobial peptides (AMPs) are universal effector molecules that directly eliminate invading pathogenic bacteria, fungi, viruses, and parasites. In mammals, including humans, such peptides may also modulate the adaptive immune systems. To date, more than 2890 AMPs have been identified in bacteria, fungi, plants, and animals. To better manage this information, my laboratory has established the Antimicrobial Peptide database (APD) as a tool for AMP naming, classification, search, statistical analysis, prediction, and design. Our database is also a useful resource for developing novel antimicrobial agents. These miniature proteins are capable of adopting a variety of three-dimensional structures, inspiring our design of natural mimics that benefit mankind. The objectives of our current NIH-funded projects are to develop AMPs into novel antimicrobial agents, including covalent immobilization on medical implants.
We are particularly interested in an in-depth understanding of the functional roles of human AMPs and their relationships with human diseases, including cancer. Recently, we have solved high-quality structures of human cathelicidin LL-37 and its important fragments by multidimensional nuclear magnetic resonance (NMR) spectroscopy. Dioctanoyl phosphatidylglycerol (D8PG) has been established as a new and unique membrane-mimetic model, which enables the detection of Phe-PG and Arg-PG interactions. Based on three-dimensional structures, we have identified the most potent region within LL-37 against MRSA, thereby identifying a useful template for designing novel therapeutic compounds against this superbug (US Patent 7,465,784). To elucidate the mechanism of action, we are utilizing a variety of biophysical and biochemical techniques. Our studies will lay the foundation for peptide engineering with a goal to overcome the hurdles (stability, toxicity, and production) on the way to the development of natural AMPs into novel therapeutics.
Another research direction of high interest to us is to engineer molecules that control protein-mediated signal transduction pathways essential for bacterial survival or infection in collaboration with colleagues in the Center for Staphylococcal Research. The lead compound will be optimized by combining NMR-based library screening with rational design based on three-dimensional structures of protein-protein complexes.

