Our Enterovirus Research Group focuses on a specific enterovirus called the group B coxsackieviruses (CVB). The coxsackieviruses were named after being originally discovered in the small Hudson River town of Coxsackie in New York in the late 1940s. CVB are human enteroviruses, a genus within the large Picornavirus family. For a look at the diversity of these viruses, go to the Picornavirus Home Page. The enteroviruses that most people know by name are the polioviruses, the viruses which used to cause annual outbreaks of paralytic poliomyelitis but which now are eradicated in our part of the world and in the process of eradication elsewhere. This wonderful improvement in our health was brought about by the development of the poliovirus vaccines.
There are 6 CVB serotypes, or CVB1-6. Serotypes mean that if a person is infected by one serotype, he or she will develop protective immunity against that one serotype. For example, if you are infected by CVB6, you will develop protective immunity against CVB6 but you will not develop at the same time protective immunity against the other CVB serotypes, 1-5. This is a practical definition of serotype: protective immunity to one type does not confer protective immunity to another type, because they are immunologically distinct. Protective immunity means that while you can actually become re-infected with the same serotype, the infection will be short lived, because you have circulating antibodies that recognize the virus and quickly neutralize it. The CVB are closely linked to the causation of several severe diseases and many less severe conditions. Among the most studied severe conditions, are inflammatory heart muscle disease or myocarditis, pancreatitis (inflammation of the exocrine or acinar portion of the pancreas, but not Islets of Langerhans) and aseptic meningitis. Type 1 (insulin dependent or juvenile) diabetes can also be triggered by a CVB infection. A good place to start looking for more information on enteroviral and CVB diseases is at the Centers for Disease Control and Prevention. The CVB are similar to other human enteroviruses. You probably have heard of the polioviruses, viruses that are the cause of poliomyelitis, a paralyzing disease that was epidemic prior to the development of the poliovirus vaccines. Poliovirus is an enterovirus. The rhinoviruses, viruses that cause the common cold, are also very similar to the CVB and are now officially characterized as enteroviruses due to this close similarity. All of these viruses are in the family of picornaviruses. You can discover more information about these and other viruses at All The Viruses on the World Wide Web.
The CVB entity is called a virion. The shell that protects the genetic stuff of the virus is the capsid in which the viral genetic stuff is found. The viral genetic stuff is called the viral genome. The viral genome is made of a single strand of positive sense RNA. Positive sense means that this RNA, when it enters the cell through the infectious process, becomes a messenger RNA that permits the cell protein synthesis machinery to read the RNA and produce the viral proteins. Once enough viral proteins are made, they copy the viral genome and eventually, new virions, stuffed with 1 strand of RNA, are assembled. This completes the infectious cycle. By this time, the cell is very sick and dies, breaking open and spilling the new viruses out into the body. This is a very simple explanation of how these viruses spread once in the body. The CVB virion is composed of 4 different proteins, called capsid proteins.
 |
 |
|
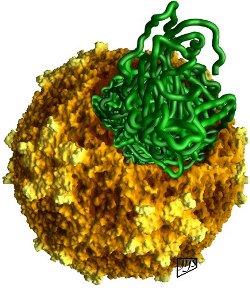
This image is of a quite different picornavirus called Mengo virus, a pathogenic virus of mice predominantly but which readily infects any mammal. The colors are, of course, from the imagination of the creator of this image, and help to define the topography of these capsid structures. This image shows the Mengo virus capsid structure (in browns and yellows) from which a section has been removed to envision how the RNA strand (in green) might appear. It gives the viewer an idea how the organization of the virion works, although this is not how the infectious process procedes in all likelihood. |
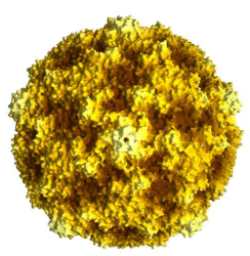
This is another computer generated image of the coxsackievirus B3 (CVB3) capsid structure. All CVB capsids, of any serotype, will look identical to this one. Facing the viewer is a center of a five-fold axis of symmetry. Look to the top of the structure and there you will also see another 5-fold center. Note how it is the highest protruding structure on the surface of the capsid. Look again at the center and then around it. You can see repeating structures, five times all around the structure. One may also draw a dividing line between the two 5-fold centers we have identified, from the middle one to the top one. If you do, that which is on the left is symmetrical to that which is on the right. This is one of the two-fold axes of symmetry on the capsid structure. There are also 3-fold axes of symmetry on the virus structure; can you find one? |
| These two images are the product of Dr. Jean-Yves Sgro at the Institute of Molecular Virology at the University of Wisconsin, Madison campus, where more of his excellent images can be seen. | |
These capsid proteins assemble to form the capsid structure, which has icosahedral symmetry. The figure here shows how the capsid probably looks if it were large enough to see. The structure was derived in the laboratory of the late Professor Michael Rossmann during his time at Purdue University. Capsid structures are often derived using a technique called x-ray crystallography, in which virus is concentrated and then crystallized, much as you might crystallize sugar or salt (although it is much more complicated than that). The crystal forms because, like salt or sugar molecules, the capsid structure of these viruses is regular and repeating in shape. Once the crystal is made, x-rays are passed through it and the crystal bends the rays. These results are captured on film and analyzed using complex mathematics, then formulated into a structure. Another highly informative website to view virus structures is at the University of Wisconsin's Institute for Molecular Virology - just click on the Images and Animation Database icon on this page to learn more.

